Comparative Analysis of 2D and 3D Differentiation Approaches for iPSC-Derived Neural Cells

Novoron Bioscience, San Diego, CA
- Matthew J. Patterson
- David A. Kochman
- Leah Boyer PhD

Defined Bioscience, San Diego, CA
- Jerome V. Karpiak PhD
The differentiation of induced pluripotent stem cells (iPSCs) into neural progenitor cells (NPCs) and neurons provides essential tools for understanding neural development, modeling diseases, and developing therapeutics. Protocols leveraging 2D monolayer cultures and 3D systems each offer distinct benefits, suiting diverse research objectives. While 2D methods are well- established for their simplicity and planar structure that has advantages for imaging, 3D approaches allow for the recapitulation of more physiologically relevant cellular microenvironments and cell types.
These differentiation protocols have proven valuable for neuroscience research and therapeutic development. They allow for the study of neural development in vitro, providing models to investigate disease mechanisms for neurodegenerative conditions such as Alzheimer’s and Parkinson’s disease. Moreover, these systems support the discovery of new therapeutics by enabling high-throughput drug screening in human-derived neural cells, as well as personalized medicine approaches using patient-specific iPSC lines. The ability to develop human NPCs and mature neurons in 2D and 3D systems is crucial for improving the translation of research findings to clinical applications.
In both protocols presented here, iPSCs were cultured using Defined Bioscience's HiDef® B8, a complete, serum-free medium designed for the feeder-free expansion of human pluripotent stem cells. HiDef-B8 ensures consistent and robust iPSC growth while maintaining pluripotency, providing a solid foundation for downstream neural differentiation.
iPSC generation
This study leverages iPSCs derived from a female donor ("iPSC-1") and an unrelated male donor ("iPSC-2") to ensure robustness and biological relevance from distinct genetic backgrounds across both sexes. iPSCs were cultured using HiDef-B8 (Defined Bioscience), a serum-free, defined medium optimized to maintain pluripotency and promote robust cell growth. These cells were expanded under feeder-free conditions, single cell-passaged with Accutase™ (Innovative Cell Technologies) or aggregate-passaged with collagenase IV. iPSC lines were verified using key pluripotency markers independent of those used for reprogramming including Nanog and TRA-1-81 (Figure 1). These karyotypically stable iPSCs serve as the foundation for downstream neural differentiation, supporting the development of both 2D and 3D neural differentiation workflows (Figure 2).
Neural Differentiation Workflows
2D Differentiation Workflow (Figure 2, Upper Panel): The 2D workflow, based on Chambers et al. (2009) and updated in Manos et al. (2022), employs a monolayer culture system starting with iPSCs grown to confluency in HiDef-B8 medium and then briefly in HiDef-B6, formulated without FGF2-G3 and TGFβ3. Neural induction is initiated by treating the cells with SMAD inhibitors (SB432542 and LDN193189), promoting a direct and efficient transition to neural progenitor stages. After replating, the cells are expanded as NPCs and then differentiated into neurons using Neurobasal™ (Thermo-Fisher) medium supplemented with B27, alanyl-glutamine, and key neurotrophic factors (BDNF, GDNF, d-cyclic AMP, ascorbic acid, and laminin) and small molecules (RO4929097, cyclopamine, LDN193189). This streamlined, high-throughput- compatible method provides a practical platform for modeling neural processes in vitro.
3D Differentiation Workflow (Figure 2, Lower Panel): The 3D differentiation approach, adapted from Boyer et al. (2012), begins with the expansion of iPSCs in HiDef-B8 medium. Colonies are transferred onto shaker flask to generate neuroectoderm embryoid bodies (nEBs) grown in suspension, mimicking in vivo-like cellular aggregates. Over the course of differentiation, nEBs are plated onto laminin-coated surfaces to form rosettes, reflective of early neural tube formation, which are manually selected and expanded to establish neural progenitor cell (NPC) lines. These NPCs can be expanded greater than ten passages, cryopreserved, and further differentiated into neurons using BrainPhys™ (STEMCELL Technologies) medium enriched with neurotrophic supplements such as BDNF, GDNF, cAMP, and ascorbic acid. This workflow recapitulates neural architecture in a three-dimensional context, offering a physiologically relevant system for studying neural development and function.
Image analysis from iPSC to Neurons
Both the 2D and 3D differentiation methods demonstrate distinct morphological transitions from iPSCs to mature neurons. In the 2D protocol (Figure 3A), iPSCs display compact colony morphology, progressing through pre-NPC and NPC stages into neuron-like cells. The 3D method (Figure 4A) begins with nEB formation from suspended iPSC colonies, followed by rosette generation, NPC expansion, and neural differentiation. nEBs and rosettes exhibit characteristic morphologies, indicative of neural lineage commitment. Across both approaches, female (iPSC-1) and male (iPSC-2) iPSC lines exhibit consistent developmental patterns, highlighting the reproducibility of these workflows.
Immunofluorescent (IF) analysis further confirms the neural differentiation achieved in both methods. In the 2D system (Figure 3B), NPCs exhibit strong SOX2 (green) and NESTIN (red) co-expression, indicating neural progenitor identity, with PAX6 (green) marking advanced neural commitment. Similarly, the 3D method (Figure 4B) shows robust expression of SOX2 and NESTIN during the NPC stage, with additional FOXG1 (green) co-expression identifying forebrain-specific NPC populations. These markers confirm the generation of stage- specific neural cells, with the 3D method offering additional complexity and regional specification.
Together, these results underscore the utility of 2D and 3D differentiation workflows in generating diverse and reproducible neural populations, suitable for a variety of research and therapeutic applications.
Image analysis of 2D and 3D Neurons
Immunofluorescent staining of neurons derived using the 2D method reveals the robust expression of neuronal markers such as TUJ1 (green) and synapsin (red), indicating well-differentiated neuronal networks. Additionally, GFAP (red), a marker of astrocytes, demonstrates the presence of glial cells alongside neurons, suggesting the development of a co-culture environment. Images from both female (iPSC-1) and male (iPSC-2) iPSC- derived neural cultures show consistent neuronal and glial marker expression, highlighting the reproducibility of the 2D method across different donor lines.
The 3D differentiation method yields neurons with distinct morphological characteristics and more intricate network formation compared to the 2D method. IF staining reveals pronounced TUJ1 (green) expression alongside synapsin (red), reflecting the formation of synaptic connections. GFAP (red) staining indicates the presence of astrocytes, and their three-dimensional arrangement mirrors the complexity of in vivo neural environments. Similar to the 2D method, the 3D protocol demonstrates consistent outcomes across female (iPSC-1) and male (iPSC-2) iPSC lines, reinforcing the reliability of this approach for generating advanced neural models.
These results highlight the capacity of both methods to generate neuron-astrocyte networks, with the 3D method offering enhanced structural complexity, suitable for modeling intricate CNS processes.
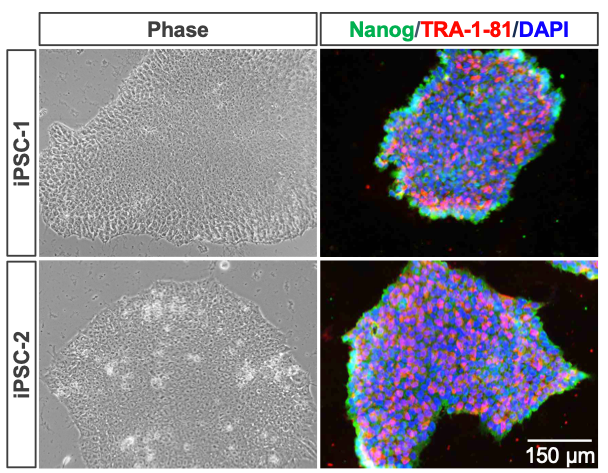
Figure 1: Development and use of iPSCs derived from male and female donors.

Figure 2. Schematic representation of the 2D, upper panel, and 3D, lower panel, neural differentiation workflows, including key media, supplements, and timelines. Ri indicates the ROCK inhibitor Y-27632.
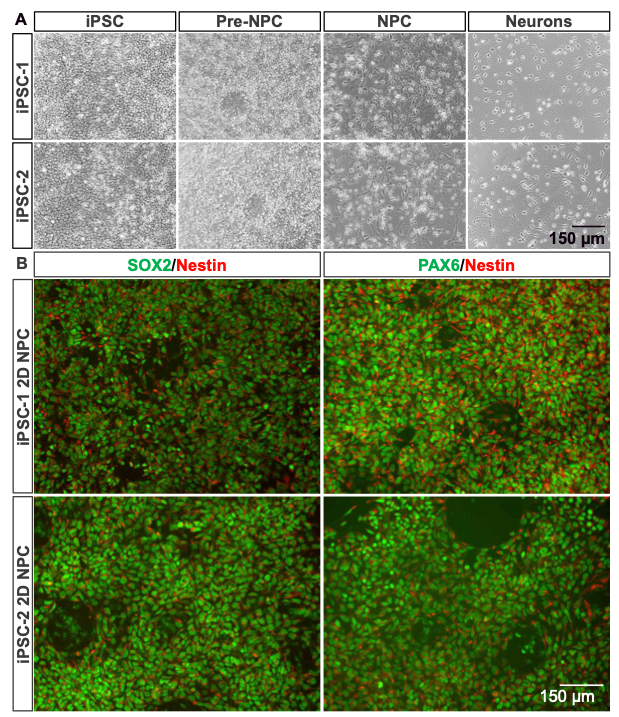
Figure 3. Phase-contrast and IF images demonstrating rosette formation, NPC maturation, and neuron differentiation using the 2D protocol.
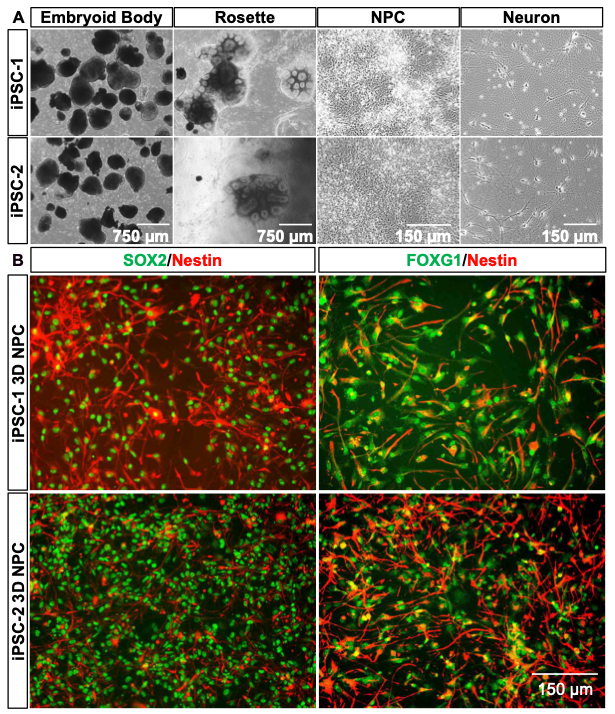
Figure 4. Phase-contrast and IF images illustrating 3D neural differentiation, including embryoid bodies, rosettes, and mature neurons.
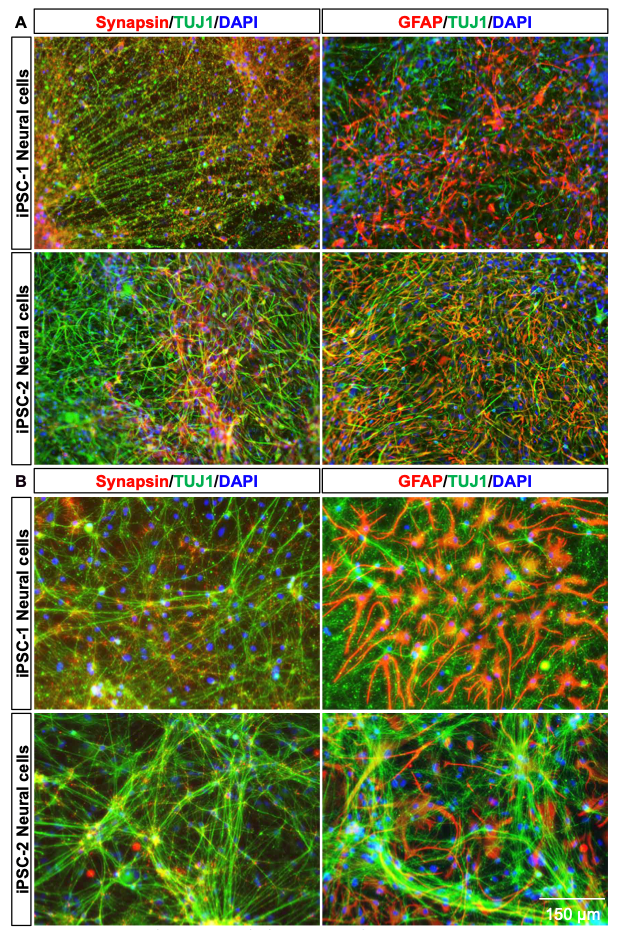
Figure 5. Immunofluorescent (IF) analysis of neuronal and glial marker expression in 2D (A) and 3D (B) neural cultures.
Both 2D and 3D differentiation approaches for iPSC-derived NPCs and neurons offer valuable tools for advancing neuroscience research and therapeutic development. The 2D method provides a streamlined platform ideal for early-stage studies and drug screening, and the 3D method offers a more physiologically relevant system for exploring complex cellular interactions and disease mechanisms that includes a highly expandable, easily banked multipotent NPC intermediate stage. Both approaches have been adapted to 384-well plates and been successful in finding drugs that are in clinical trials.
The application of these protocols is critical for addressing significant unmet needs in the treatment of central nervous system (CNS) disorders. Novoron Bioscience leverages these differentiation assays in their innovative research programs aimed at developing therapies for neurological damage and disease. Their work spans a variety of conditions, including tauopathies, such as Alzheimer’s disease, as well as overcoming the inhibitory effects of myelin seen in multiple sclerosis and spinal cord injuries. By incorporating robust iPSC- based systems, Novoron aims to translate these models into actionable insights that drive therapeutic breakthroughs.
Defined Bioscience’s HiDef-B8 medium and reagents plays an important role in the success of these workflows, ensuring consistent and reproducible iPSC expansion as a foundation for these downstream applications. This collaboration highlights the utility of iPSC technology in addressing some of the unmet needs in neuroscience. Contact Defined Bioscience to learn about related HiDef-B6 medium, HiDef-N2 neural supplement, thermostable FGF2-G3, and Ready-CEPT cocktail to support clonal robustness in single-cell expansion and reduce selection pressure for high passage and suspension PSC workflows.
If you're interested in learning more, we’d love to hear from you.
Boyer, L. F., Campbell, B., Larkin, S., Mu, Y., & Gage, F. H. (2012). Dopaminergic Differentiation of Human Pluripotent Cells. Current Protocols in Stem Cell Biology, 22(1). https://doi.org/10.1002/9780470151808.sc01h06s22
Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M., & Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnology, 27(3), 275–280. https://doi.org/10.1038/nbt.1529
Manos, J. D., Preiss, C. N., Venkat, N., Tamm, J., Reinhardt, P., Kwon, T., Wu, J., Winter, A. D., Jahn, T. R., Kiran Yanamandra, Titterton, K., Karran, E., & Langlois, X. (2021). Uncovering specificity of endogenous TAU aggregation in a human iPSC- neuron TAU seeding model. Science, 25(1), 103658–103658. https://doi.org/10.1016/j.isci.2021.103658
Neurobasal™, BrainPhys™ and Accutase™ are trademarks of Thermo-Fisher, STEMELL Technologies, and Innovative Cell Technologies, respectively.

HiDef® B8 Stem Cell Growth Medium
HiDef® B8 Stem Cell Growth Medium
Choose your option

DMEM/F12 Basal Medium
DMEM/F12 Basal Medium
Choose your option
Ready-CEPT® Cell Viability Enhancer
Ready-CEPT® Cell Viability Enhancer
Choose your option

FGF2-G3 (bovine) growth factor
FGF2-G3 (bovine) growth factor
Choose your option